Research
Modeling of amino acid transport across cell membrane
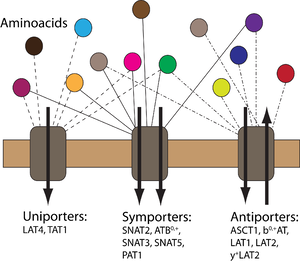
Homeostatic regulation of amino acids (AA) is complex and involves distribution of AAs between cells and tissues with numerous translocation steps across cellular membranes via transport proteins. AAs are taken up into epithelial cells via apical secondary active transporters from the lumen of small intestine and kidney proximal tubule and subsequently released in the extracellular fluid via another set of basolateral transporters. The interaction of several transporters (uniporters and antiporters) makes the AA transport dynamics rather complicated. In this project, we aim to develop a computational model to represent the AA transport dynamics across cell membranes. In collaboration with the Epithelial Transport Group at the Institute of Physiology, University of Zurich, experiments will be performed on a Xenopus laevis oocyte expression system both for endogenously and exogenously expressed transporters. Experimental results will be used to characterize first the Xenopus laevis oocyte expression system with the endogenously expressed transporters and then to characterize each exogenously expressed transporter-AA couple through the use of computational modeling. The ultimate goal is to create a simulation environment for the transport of AAs through cell membranes.
Related publications
- Analysis of L-leucine amino acid transporter species activity and gene expression by human blood brain barrier hCMEC/D3 model reveal potential LAT1, LAT4, B0AT2 and y+LAT1 functional cooperation Journal of Cerebral Blood Flow & Metabolism, 42(1), 90 – 103 (2022).
- Functional polarity of microvascular brain endothelial cells supported by neurovascular unit computational model of large neutral amino acid homeostasis Frontiers in Physiology, 9(171), 1 – 14 (2018).
- Quantifying the relative contributions of different solute carriers to aggregate substrate transport Scientific Reports, 7(40628), 1 – 13 (2017).