Studienarbeiten
Real-time integration of clinical measurements and computational models in personalised medicine: enhancing the power of non-invasive diagnostic techniques
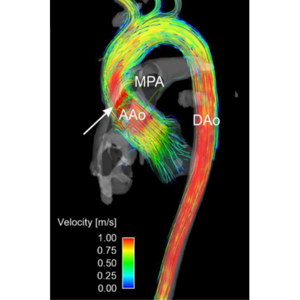
Non-invasive imaging methodologies, such as Magnetic Resonance Imaging (MRI) and Computed Tomography (CT) are often used in clinical routine to acquire patient specific data for diagnostic purposes. Phase contrast MRI (PC-MRI) can be used, for example, to visualise blood flow in large vessels, such as the renal or iliac artery. Poor resolution and substantial noise are the main drawbacks of this technique, requiring post-processing methodologies to enhance the measurements.
Improving these two factors may enhance the diagnostic power of MRI, providing the clinician with important information about non-physiological flow structures that might be correlated with pathologies. Proper orthogonal decomposition (POD) of computational fluid dynamics (CFD) simulations has been proposed as a means to improve PC-MRI acquisitions [1]. However, the feasibility and benefit of this approach when applied to 3D patient-specific cases remains to be ascertained.
In this project, the student will extend the methodology detailed in [1] to enhance 3D PC-MRI in the clinical routine. For the scope of this project, we will focus on the reconstruction of blood flows at the bifurcation between the descending aorta and renal artery. The envisioned workflow will include the characterisation of important geometrical and fluid-dynamic parameters of renal arteries, generation of a dataset of realistic hemodynamic simulations, and finally the reconstruction of any new hemodynamic configuration from this pre-computed dataset using POD decomposition. Attention will be paid to the number of data points required to fully reconstruct the flow field in a new geometry and reduce noise. After validation in synthetic cases, the student will apply his methodology to patient-specific PC-MRI datasets. The thus-reconstructed high-resolution patient-specific flow-field will allow for the analysis of hemodynamic parameters of pathophysiological interest, such as regions of flow recirculation or flow separation, pressure drops or shear stresses.
In this project, the student:
- will benefit from the collaboration with an interdisciplinary group with competences going from aerospace engineering to virtual reality to medicine and biology,
- work with real anatomic data and state-of-the-art numerical models,
- contribute to an existing framework for personalised medicine.
Prerequisites:
- basic understanding of computational fluid dynamics,
- basic programming knowledge,
- strong motivation and willingness
References:
[1] McGregor et al (2008), Exploring the Use of Proper Orthogonal Decomposition for Enhancing Blood Flow Images Via Computational Fluid Dynamics, Medical Image Computing and Computer-Assisted Intervention – MICCAI 2008, 5242:782-789. Link.
[2] Stankovic et al (2014), 4D flow imaging with MRI , Cardiovascular Diagnosis & Therapy, 4(2):173-192. Link.
Information
(position closed)
For further information, please contact
Prof. Vartan Kurtcuoglu
Dr. Stefano Buoso
Dr. Diane de Zélicourt